Shimadzu has produced an application note introducing a new and valuable angle to the evaluation of bioprocess by elucidating the state of metabolic pathway in which the target end product is involved.
 Introduction
Introduction
Throughout the long history of fermentation and bioengineering, conditions that affect microbial growth have been continuously optimized to maximize the yield and quality of end product, from alcoholic beverages to biofuel production. In recent years, this is performed systematically by monitoring relevant parameters and applying the disciplines of engineering, which is a growing field of investigation known as bioprocess engineering.
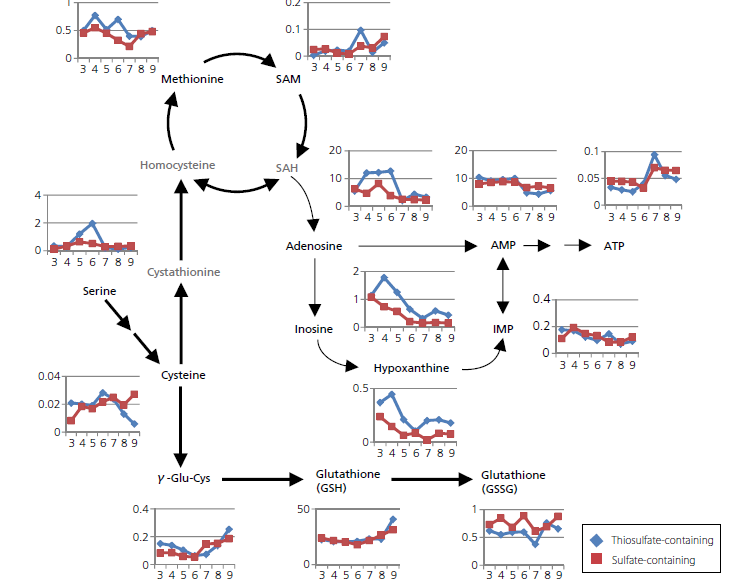
In this application note metabolomic analysis of E.coli extracts have been performed by fast, label-free methods using LC-MS/MS (supplied as the Method Package for Primary Metabolites), which were able to simultaneously monitor the levels of sulfur-containing metabolites, such as cysteine, glutathione, methionine, as well as other metabolites downstream and upstream of the pathway.
Method
E. coli was cultured in minimal media to which thiosulfuric acid or sulfuric acid was added as a sulfur
source. To evaluate metabolic changes during culture, some E. coli were collected from the culture suspension at 3, 4, 5, 6, 7, 8, and 9 hours of culture. The optical density (OD) of the collected E. coli was measured before the media components and E. coli were quickly separated by filtration. E. coli extract was then prepared by breaking down the isolated E. coli in methanol. After removing methanol by centrifugal concentration, the extract was adjusted to an appropriate dilution with ultrapure water and used for LC-MS analysis. Metabolites were analysed simultaneously using the analytical conditions of an ion
pairing method (LCMS-8040) and non-ion pairing method (LCMS-8050) obtained from an LC-MS method
package [primary metabolites].
 Results
Results
In this application note it was observed how metabolites change during the course of E. coli culture, focusing on sulfur containing metabolites that are linked to cysteine production. The time-course changes in the abundance of metabolites were compared between the same bacterial strain cultured with media supplemented with either thiosulfate or sulfate. The results highlighted how different sectors of the metabolic pathway were affected by respective supplementation of ingredient. This approach is expected to facilitate the designing of bioprocess or manipulation of strain.




