Recently a reader sent me a question regarding a problem he was having with recovery of a vitamin he was measuring in pet foods. He found that the precision, measured as repeatability of peak area for a given sample, was adequate for multiple products, but the assay amount was low by 10–40%, depending on the product. After further probing of the conditions, I found that he was running a calibration curve based on reference standards dissolved in an aqueous solution, instead of spiked into blank sample matrix. I feel that this is the basis of the problem.
Matrix Effects and Recovery
The sample matrix, whether it is dog food, blood plasma, sewage sludge, or some other material, can have a profound impact on the analytical results for an HPLC method. As a result of this, a matrix-based calibration curve is recommended almost universally. In many methods, I consider working out the sample cleanup procedure to be the most difficult part of method development. There are several goals of sample preparation. One is to remove materials that might interfere chromatographically with the analyte, another is to get acceptable recovery of the analyte, and a third is to remove “column killers” – those matrix component that shorten column lifetime. A common way to test recovery is to spike known amounts of reference standard into a blank matrix, or placebo, mix it well, then perform sample extraction. The recovered amount then can be compared to the amount spiked into the matrix to determine recovery. This is done by comparing the signal of the extracted sample to the signal obtained from pure reference standard. Although it is ideal to get 100% recovery of the analyte, this seldom occurs. In many cases, recovery of 80-95% can be obtained, but in others, recovery may be below 50%. Although the method usually will perform more reliably and reproducibly the higher the recovery, this is not necessarily the case or a requirement for a successful method.
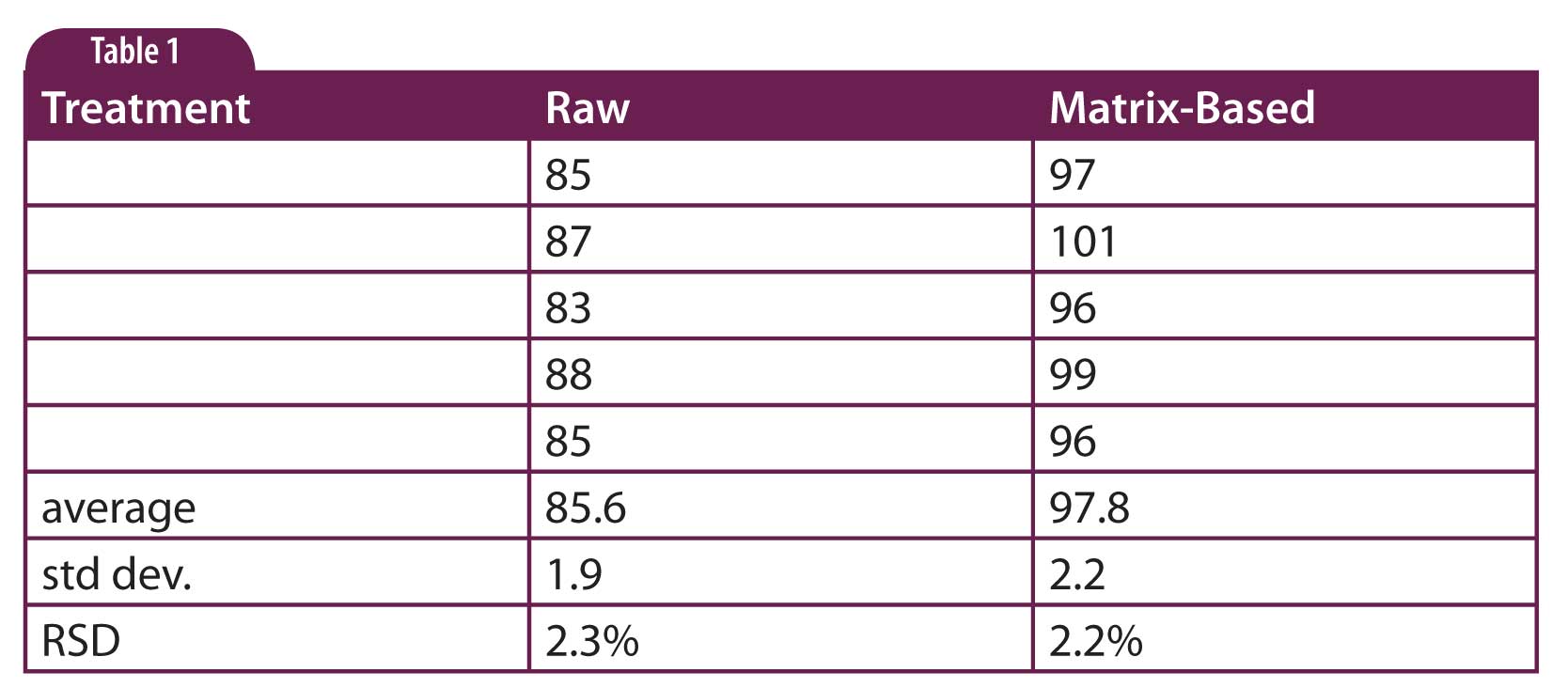
Table 1
Matrix-Based Calibrators
If, as in Table 1 (column labeled “raw” data), you can consistently recover 86% of the sample that you spike into a blank matrix, it is expected that you will also recover 86% of the analyte from a real sample. Then the extracted samples can be directly related to the true amount of analyte present. The simplest way to do this is to use a matrix-based calibration curve. This is done by spiking different concentrations of reference standard into blank matrix, extracting the standard back out, and using the resulting solutions to construct the calibration curve. This will correct for the low recovery from the sample matrix of both real samples and the calibrators so that the reported values should be close to the real values (Table 1, “matrix-based” data). Because the matrix can have such a profound influence on the final results, it is recommended to match the matrix for the calibration curve to the sample. That is if you are analyzing vitamin D in dry cat food, you need to use untreated dry cat food as the matrix, not dry dog food. Of course, it isn’t always possible to make an exact match of the matrix. For example, when analyzing human plasma samples for a drug used in a clinical trial, a single matrix-based calibration curve is used for multiple subjects, so small differences between subjects are not compensated for. In some cases, the calibration curve needs to be based on pre-dose plasma from that individual, but the need for this level of correction is rare.
Sometimes you can get away without using a matrix-based calibration curve, but this should be done only when you have shown that there is no difference in the analytical results with and without the matrix present. Finally, your industry may have standard practices that differ from the recommendations made here, so you should go with the best practices for your particular application.
This blog article series is produced in collaboration with John Dolan, best known as one of the world’s foremost HPLC troubleshooting authorities. He is also known for his research with Lloyd Snyder, which resulted in more than 100 technical publications and three books. If you have any questions about this article send them to TechTips@sepscience.com




